Getting Started: Creating Your First Montage Mosaic
Montage is a command-line toolkit containing modules for reading a collection of images and performing all the processing steps needed to assemble them into a mosaic.
You build mosaics by running processes in sequence at the command line, or automating them in scripts. We recommend a new user step through the commands once to become familiar with their operation, and then use scripts to automate the process.
Let's Build A Mosaic!
We will assume that you have built the current version of Montage on a Unix or Linux platform, that you have added Montage to your path (very important!), and are ready to run jobs from the command line.
We will perform the simplest example of building a mosaic of a set of images on your desktop, keeping the spatial sampling, coordinate system and projection of the original images. We'll build a 1.5 deg x 1.5 deg mosaic of 91 2MASS K-band images centered on M17. These data were downloaded from the NASA IPAC Infrared Science Archive (IRSA). The duration for each step indicates the time taken on a Mac Pro machine with a 3.2 GHz Quad-Core Intel Xeon processor and 16 GB 1066 MHz DDR3 memory, running Mac OS 10.9.5.
Preparation
Create a directory called Ktutorial and cd into it:
- cd Ktutorial
Download the 2MASS images (144 MB) and uncompress the file with these commands:
- $ gunzip Kimages.tar.gz
- $ tar xvf Kimages.tar
The input images will be placed in a subdirectory called Kimages.
Create the following additional subdirectories under Ktutorial. They will help manage all the intermediate files we will create at each step.
Ktutorial
|--Kprojdir/
|--diffdir/
|--corrdir/Building the mosaic:
Part One: Build a Mosaic Without any Background Corrections
1. Create a metadata table for the input images: (2 seconds)
- mImgtbl Kimages Kimages.tbl
- [struct stat="OK", module="mImgtbl", count=91, nfile=91, nhdu=91, badfits=0, badwcs=0]
Where:
| Kimages | Name of the directory containing the images to be reprojected |
| Kimages.tbl | Output image metadata table |
2. Process the image metadata table to create a header file, Ktemplate.hdr, that describes the footprint of the mosaic on the sky in FITS/WCS convention. This header will become the FITS header of the final mosaic. (1 second)
- mMakeHdr Kimages.tbl Ktemplate.hdr
- [struct stat="OK", module="mMakeHdr", msg="Cube columns exist but are either blank or inconsistent. Outputting 2D only.", count=91, ncube=0, naxis1=6110, naxis2=6857, clon=275.185261, clat=-16.248875, lonsize=1.697222, latsize=1.904722, posang=359.953803, lon1=276.072612, lat1=-17.199892, lon2=274.296309, lat2=-17.198517, lon3=274.306471, lat3=-15.294203, lon4=276.065648, lat4=-15.295564]
where
| Kimages.tbl | Input image metadata table |
| Ktemplate.hdr | Output mosaic header file |
Here are the contents of Ktemplate.hdr:
SIMPLE = T
BITPIX = -64
NAXIS = 2
NAXIS1 = 6110
NAXIS2 = 6857
CTYPE1 = 'RA---TAN'
CTYPE2 = 'DEC--TAN'
EQUINOX = 2000
CRVAL1 = 275.185260873
CRVAL2 = -16.248875428
CRPIX1 = 3055.5000
CRPIX2 = 3429.0000
CDELT1 = -0.000277778
CDELT2 = 0.000277778
CROTA2 = 359.953802926
END
3. Reproject the input images to the specifications for the mosaic (34 seconds):
- mProjExec -p Kimages Kimages.tbl Ktemplate.hdr Kprojdir Kstats.tbl
- [struct stat="OK", module="mProjExec", count=91, failed=0, nooverlap=0]
where:
| Kimages | Name of the directory containing the images to be reprojected |
| Kimages.tbl | Image metadata table describing the files |
| Ktemplate.hdr | Header template for the mosaic |
| Kprojdir | Directory into which reprojected images will be written |
| stats.tbl | Name of the output table containing processing times and status for each image |
4. Create a new metadata table that contains the updated header information in the reprojected files (1 second):
- mImgtbl Kprojdir/ images.tbl
- [struct stat="OK", module="mImgtbl", count=91, nfile=91, nhdu=91, badfits=0, badwcs=0]
where
| Kprojdir | Name of the directory containing the reprojected images |
| images.tbl | Output image metadata table |
5. Create a mosaic without background corrections (17 seconds):
- mAdd -p Kprojdir/ images.tbl Ktemplate.hdr m17_uncorrected.fits
- [struct stat="OK", module="mAdd", time=12]
where
| Kprojdir | Name of the directory containing the reprojected images |
| images.tbl | Output image metadata table |
| Ktemplate.hdr | Header template for the mosaic |
| m17_uncorrected.fits | Name of the output file to be created |
6. Create a PNG file of the mosaic for visualization (24 seconds):
- mViewer -ct 1 -gray m17_uncorrected.fits -1s max gaussian-log -out m17_uncorrected.png
- [struct stat="OK", module="mViewer", type="grayscale", width=6101, height=6857, min=632.408, minpercent=9.66, minsigma=-1.00, max=10484, maxpercent=100.00, maxsigma=728.38, datamin=299.523, datamax=10484, xflip=0, yflip=1, bunit="", colortable=1]
The PNG file looks like this:
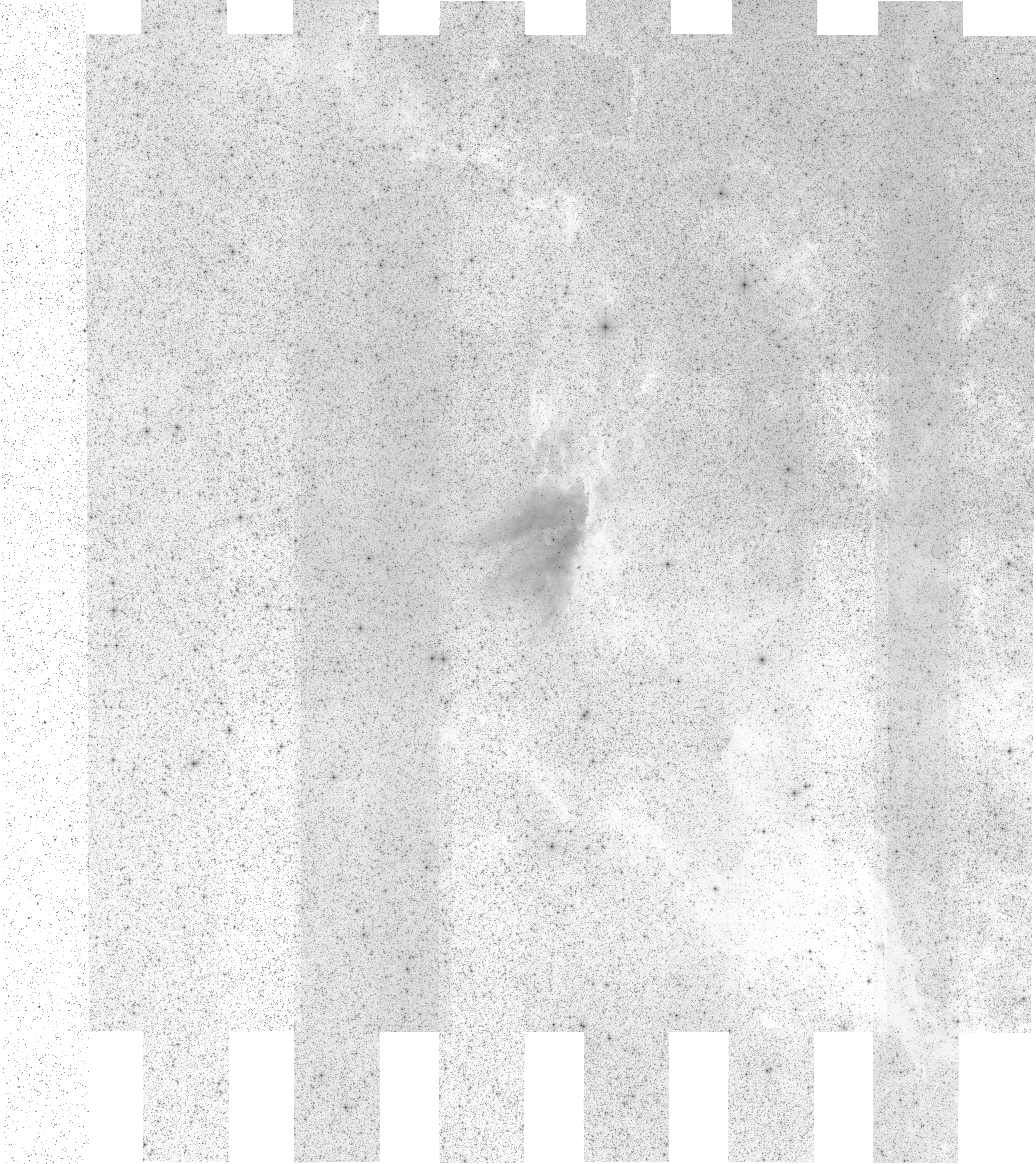
The vertical stripes represent differences in backgrounds between the input images. Let's go ahead and correct the images for these differences.
Part Two: Including Background Corrections
To smooth out the background levels between overlapping images, Montage creates a set of difference images from each pair of overlaps and fits a plane to each one.
1. Find out which images overlap (1 second):
- mOverlaps images.tbl diffs.tbl
- [struct stat="OK", module="mOverlaps", count=233]
where:
| images.tbl | Image metadata table describing the files |
| diffs.tbl | Name of the output table to be generated |
2. Subtract each pair of overlapping images and create a set of difference images in the diffdr subdirectory (13 seconds).
- mDiffExec -p Kprojdir/ diffs.tbl Ktemplate.hdr diffdir
- [struct stat="OK", module="mDiffExec", count=233, failed=0]
where:
| Kprojdir | Directory containing the reprojected images |
| diffs.tbl | Name of the differences table generated by mOverlaps |
| Ktemplate.hdr | Header template for the mosaic |
| diffdr | Directory into which the difference images will be written |
3. Calculate plane-fitting coefficients for each difference image (13 seconds):
- mFitExec diffs.tbl fits.tbl diffdir
- [struct stat="OK", module="mFitExec", count=233, failed=0, warning=0, missing=0]
where:
| diffs.tbl | Name of the output table to be generated |
| fits.tbl | Plane-fitting table generated by mFitExec |
| corrections.tbl | Global corrections table to be generated |
4. Create a table of corrections that needs to be applied to each image (1 second).
- mBgModel images.tbl fits.tbl corrections.tbl
- [struct stat="OK", module="mBgModel"]
where:
| images.tbl | Image metadata table containing information on the reprojected images |
| fits.tbl | Plane-fitting table generated by mFitExec |
| corrections.tbl | Global corrections table to be generated |
5. Apply the background removal to the original reprojected images (14 seconds).
- mBgExec -p Kprojdir/ images.tbl corrections.tbl corrdir
- [struct stat="OK", module="mBgExec", count=91, nocorrection=0, failed=0]
where:
| Kprojdir | Directory containing the reprojected images |
| images.tbl | Image metadata table describing the reprojected images |
| corrections.tbl | Set of corrections generated by mBgModel |
| corrdir | Directory into which the corrected images will be placed |
6. Create the reprojected, smoothed images into your mosaic (14 seconds):
- mAdd -p corrdir/ images.tbl Ktemplate.hdr m17.fits
- [struct stat="OK", module="mAdd", time=13]
where:
| Kprojdir | Directory containing the reprojected images |
| images.tbl | Image metadata table describing the reprojected images |
| Ktemplate.hdr | Header template for the mosaic |
| m17.fits | Name of the output file to be created |
7. Create a PNG of the mosaic for visualization (28 seconds):
- mViewer -ct 1 -gray m17.fits -1s max gaussian-log -out m17.png
- [struct stat="OK", module="mViewer", type="grayscale", width=6101, height=6857, min=632.323, minpercent=0.63, minsigma=-1.00, max=10474, maxpercent=100.00, maxsigma=889.27, datamin=291.79, datamax=10474, xflip=0, yflip=1, bunit="", colortable=1]
Here is the final mosaic (click on it to see the image a full resolution):
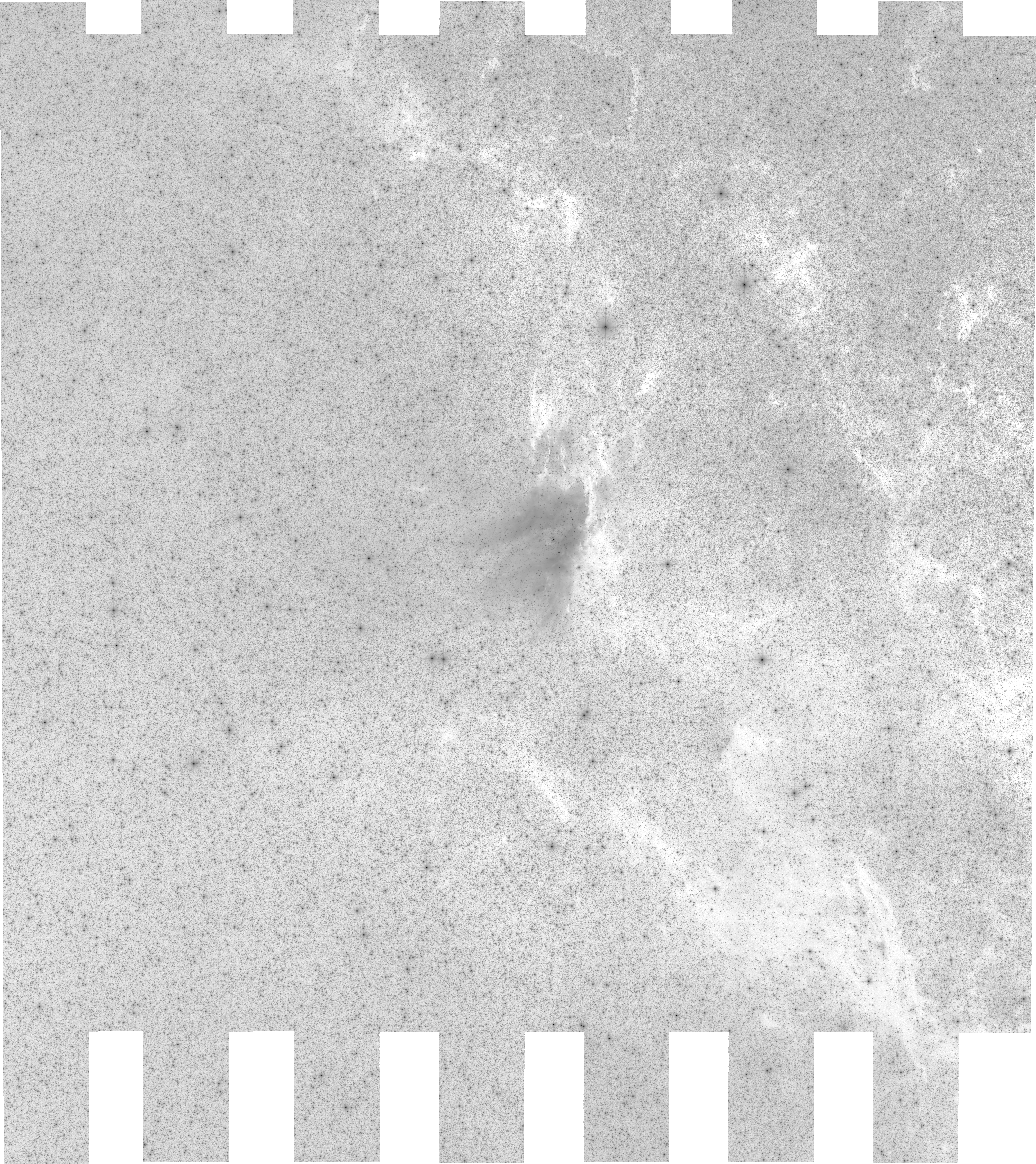
Automating the Process
Here is a simple Bash script you can run to automate the above process:
#!/bin/bash
# Mosaic of M17, K band, 1.5 deg x 1.5 deg
# Bruce Berriman, February, 2016
echo "create directories to hold processed images"
mkdir Kprojdir diffdir corrdir
echo "Create a metadata table of the input images, Kimages.tbl"
mImgtbl Kimages Kimages.tbl
echo "Create a FITS header describing the footprint of the mosaic"
mMakeHdr Kimages.tbl Ktemplate.hdr
echo "Reproject the input images"
mProjExec -p Kimages Kimages.tbl Ktemplate.hdr Kprojdir Kstats.tbl
echo "Create a metadata table of the reprojected images"
mImgtbl Kprojdir/ images.tbl
echo "Coadd the images to create a mosaic without background corrections"
mAdd -p Kprojdir/ images.tbl Ktemplate.hdr m17_uncorrected.fits
echo "Make a PNG of the mosaic for visualization"
mViewer -ct 1 -gray m17_uncorrected.fits -1s max gaussian-log -out m17_uncorrected.png
echo "Analyze the overlaps between images"
mOverlaps images.tbl diffs.tbl
mDiffExec -p Kprojdir/ diffs.tbl Ktemplate.hdr diffdir
mFitExec diffs.tbl fits.tbl diffdir
echo "Perform background modeling and compute corrections for each image"
mBgModel images.tbl fits.tbl corrections.tbl
echo "Apply corrections to each image"
mBgExec -p Kprojdir/ images.tbl corrections.tbl corrdir
echo "Coadd the images to create a mosaic with background corrections"
mAdd -p corrdir/ images.tbl Ktemplate.hdr m17.fits
echo "Make a PNG of the corrected mosaic for visualization"
mViewer -ct 1 -gray m17.fits -1s max gaussian-log -out m17.png

